Molecular Modeling Studies
Another part of our research consists in the study of the mechanism of action of the enzymes and the binding mode of the inhibitors.
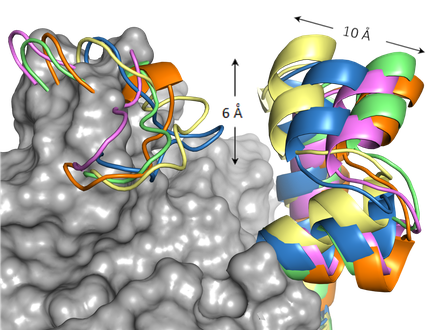
By Molecular dynamics simulation studies, we have studied the role of key residues in the enzymatic reaction of the type II dehydroquinase enzyme, an arginine and a tyrosine. Both residues reside in a flexible loop that closes over the active site upon substrate binding. These studies suggest that the essential arginine of the loop is presumed to orient the tyrosine in an appropriate manner for proton abstraction. Moreover, the aromatic ring of the inhibitors prevents appropriate orientation of the catalytic tyrosine of the loop for proton abstraction and disrupts its basicity.

MD simulation studies have allowed us to know the key interactions that control the unusual conformation required by shikimic acid in the M.
tuberculosis shikimate kinase active site and the enzyme movements that are essential for catalytic turnover.